In this project, I will perform data analysis in a dataset about smartwatches, that recoleted information like Steps, Heart Rate, Device and Gender among others. This dataset is composed of more than 6000 people, the data is available on Harvard.
In this study, researchers wanted to see if popular wearable devices, like the Apple Watch and Fitbit, could accurately predict different activities like lying down, sitting, and various levels of physical activity in a controlled lab setting. They recruited 46 participants to wear these devices while going through a 65-minute protocol, which included treadmill walking and periods of sitting or lying down. They used a method called “indirect calorimetry” to measure energy expenditure.
The researchers focused on the data collected by the devices, such as heart rate, steps, distance, and calories burned, and used machine learning models to analyze it. They tested different types of machine learning models and found that a particular model called “rotation forest” gave the best results. The accuracy of predictions was around 82.6% for the Apple Watch and 89.3% for the Fitbit.
In conclusion, this study showed that commercial wearable devices like the Apple Watch and Fitbit can reasonably predict different types of physical activities. The findings suggest that using minute-by-minute data from these devices with machine learning approaches can be a scalable way to classify physical activity types in larger populations.
Disclaimer This project involves the analysis of health data from smartwatches, sourced from a Harvard dataset. The conclusions are for educational purposes only and not medical advice. I am not a medical professional. Please consult a medical provider for personalized recommendations.
Dataset Introduction
Data Source
The data used in this project is from the paper titled “Replication Data for: Using machine learning methods to predict physical activity types with Apple Watch and Fitbit data using indirect calorimetry as the criterion” authored by Daniel Fuller. The paper’s dataset can be found on Harvard Dataverse.
- Paper Title: Replication Data for: Using machine learning methods to predict physical activity types with Apple Watch and Fitbit data using indirect calorimetry as the criterion
- Dataset: Harvard Dataverse - Replication Data for: Using machine learning methods to predict physical activity types with Apple Watch and Fitbit data using indirect calorimetry as the criterion
- Author: Daniel Fuller
- Year: 2020
- Version: V1
Importing
Importing libraries
Now we will import all the neccesary libraries that we will use for the project.
import pandas as pd
import numpy as np
import plotly.express as px
import missingno as msno
import matplotlib.pyplot as plt
Apart for this libraries that will be mainly used for representation and data handling, we will also need some libraries for the model creation, that we will use at the end of the porject.
from sklearn.model_selection import train_test_split
from sklearn.svm import SVR
Importing Data
In the offical research we can find three datasets, one with all the data combined, and the the other two separating apple watch and fitbit infromation.
health = pd.read_csv('aw_fb_data.csv', index_col = 0)
health.head()
| age | gender | height | weight | steps | heart_rate | calories | distance | resting_heart | device | activity | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 20 | Male | 168 | 65.4 | 10.7714 | 78.5313 | 0.344533 | 0.00832686 | 59 | apple watch | Lying |
| 2 | 20 | Male | 168 | 65.4 | 11.4753 | 78.4534 | 3.28763 | 0.00889635 | 59 | apple watch | Lying |
| 3 | 20 | Male | 168 | 65.4 | 12.1792 | 78.5408 | 9.484 | 0.00946584 | 59 | apple watch | Lying |
| 4 | 20 | Male | 168 | 65.4 | 12.8831 | 78.6283 | 10.1546 | 0.0100353 | 59 | apple watch | Lying |
| 5 | 20 | Male | 168 | 65.4 | 13.587 | 78.7157 | 10.8251 | 0.0106048 | 59 | apple watch | Lying |
Cleaning Data
Now we are going to clean the data from the Dataframe, this is a multiple stage proccess, we wil need to check the mssing values first, the text values and categorical values, and then the uniformity of the data.
Missing Values
To check the missing values, first we are going to check the amount of null values in the dataframe, and then we will be able to choose a path to proceed, also we can use the package missingno to visualize null values.
health.isnull().sum()
| Data | Null | Data | Null | |
|---|---|---|---|---|
| X1 | 0 | age | 0 | |
| gender | 0 | height | 0 | |
| weight | 0 | steps | 0 | |
| heart_rate | 0 | calories | 0 | |
| distance | 0 | entropy_heart | 0 | |
| entropy_setps | 0 | resting_heart | 0 | |
| corr_heart_steps | 0 | norm_heart | 0 | |
| activity | 0 | sd_norm_heart | 0 | |
| steps_times_distance | 0 | device | 0 |
In this case we dont have missing values in the dataset, that means that we can continue with the rest of the proccess, as a side note, a useful package to use to check for missing values in a visual way is to use the missingno package.
As we saw in the previous method, there is no null values in the dataframe.
Value Consistency
In this case we will check two of the columns available device and activity, the things to check are capitalization and potential blank spaces in the data inputs
device = health['device']
device.value_counts()
| Device | Count |
|---|---|
| Apple Watch | 3656 |
| Fitbit | 2608 |
activity = health['activity']
activity.value_counts()
| Activity | Count |
|---|---|
| Lying | 1379 |
| Running 7 METs | 1114 |
| Running 5 METs | 1002 |
| Running 3 METs | 950 |
| Sitting | 930 |
| Self Pace Walk | 889 |
We can see by the prevous results that there is not errors in value consistency, no need to strip blank spaces or change the capitalization.
Data Type
In all the columns we have different types of data, from categorical, to integers and floats among other, so we will check if the type is correct.
health.dtypes
| Data | Type | Data | Type | |
|---|---|---|---|---|
| X1 | int64 | age | int64 | |
| gender | int64 | height | float64 | |
| weight | float64 | steps | float64 | |
| heart_rate | float64 | calories | float64 | |
| distance | float64 | entropy_heart | float64 | |
| entropy_setps | float64 | resting_heart | float64 | |
| corr_heart_steps | float64 | norm_heart | float64 | |
| activity | object | sd_norm_heart | float64 | |
| steps_times_distance | float64 | device | object |
In this case we would not need to change any data type, except the gender, even though we will change it for names to make easier to read, for now, let’s convert it to categorical data. First we are going to see if there is any error in that category by having numbers outside the boundaries, that correspond to the part of the cleaning proccess known as insconsistent data.
health.gender.value_counts()
| Gender | Count |
|---|---|
| 0 | 3279 |
| 1 | 2985 |
We do not have any error in the data inputs, so lets convert it to categorial data.
health.gender = health.gender.astype('category')
Outliers
In this case, with health data, is more difficult to evaluate the outliers unless is a really extreme value as we do not know the premises of the study, one person can be investing a lot of time for walking for example, other data we can check that is easier to detect potential outliers can be the heart rate.
health[['hear_rate', 'height','weight', 'resting_heart']].describe()
| hear_rate | height | weight | resting_heart | |
|---|---|---|---|---|
| count | 6264 | 6264 | 6264 | 6264 |
| mean | 86.1423 | 169.709 | 69.6145 | 65.8699 |
| std | 28.6484 | 10.3247 | 13.4519 | 21.203 |
| min | 2.22222 | 143 | 43 | 3 |
| 25% | 75.5981 | 160 | 60 | 58.1343 |
| 50% | 77.2677 | 168 | 68 | 75 |
| 75% | 95.6691 | 180 | 77.3 | 76.1387 |
| max | 194.333 | 191 | 115 | 155 |
Let’s analyze the data:
Heart Rate: For this category we can observe that most of the categories are logical, except the minimum heart rate, thats 2.22.
Resting Heart: Similar to the previous case the minimum is too low, indicating a potential error, and also the maximum value is too high.
Most of the information that we can check is related to the heart rate, as thats the easier to see potential problems, the other ones, as I indicated previously, like the steps, calories or distance.
Another heart measure that can be important is the entropy for heart, as that’s used in relation to the heart variability.
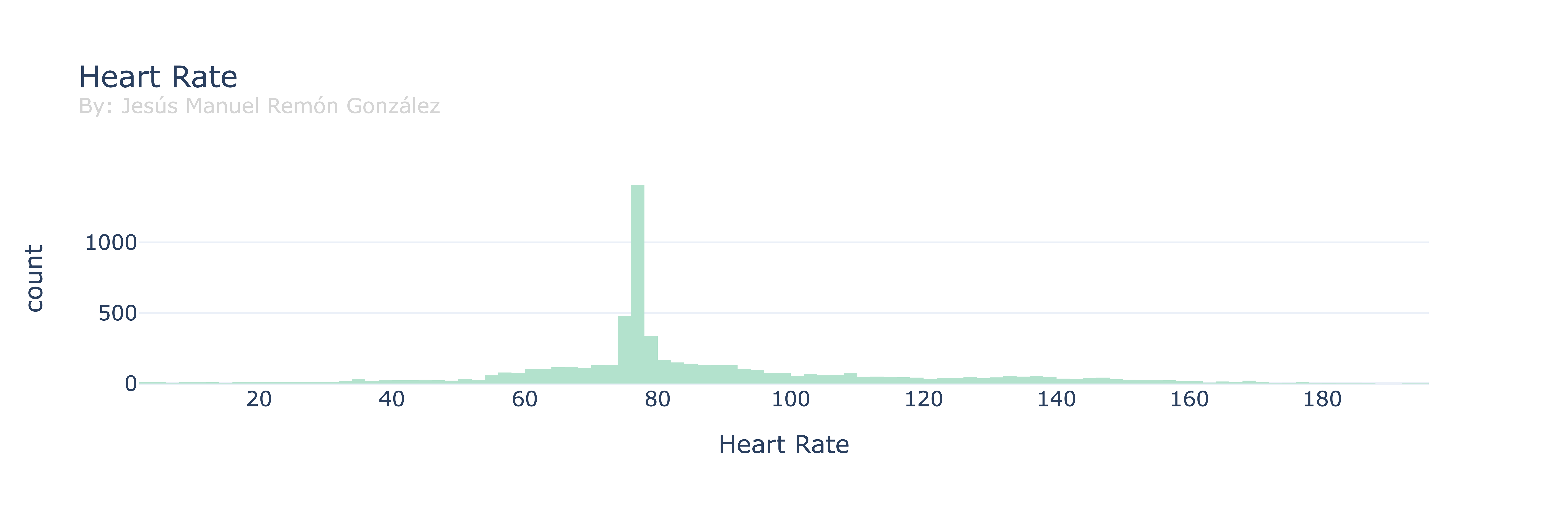
We can see a clear mode in the graphs, also we can see really lows heart rates, now we can check those cases apart, but lets extract them form the total dataset. We are going to consider as valid a low heart rate, as we do not have information if any of the components of the group have Bradycardia; according to this article form Mayo Clinic that having a heart rate of 40 bpms is not a reason to worry.
Bradycardia: Bradycardia (brad-e-KAHR-dee-uh) is a slow heart rate. The hearts of adults at rest usually beat between 60 and 100 times a minute. If you have bradycardia, your heart beats fewer than 60 times a minute. A slow heart rate isn’t always a concern. For example, a resting heart rate between 40 and 60 beats a minute is quite common during sleep and in some people, particularly healthy young adults and trained athletes.
We will now extract the rows below the 40 bpms.
lower_data = health[health['hear_rate'] <= 40.0]
We can see that there is 237 rows where the heart_rate is below the minimum, now we will eliminate that ones.
checker = health['hear_rate'] <= 40.0
health = health[~checker]
We can use an assert to see if the minimum value if above the boundarie, not getting any response means that we do not have any that is not above the limit.
assert health.hear_rate.min() > 40
After extracting the outliers from the main dataframe, we can take a look at the distribution for heart rate and resting heart rate for the outliers.
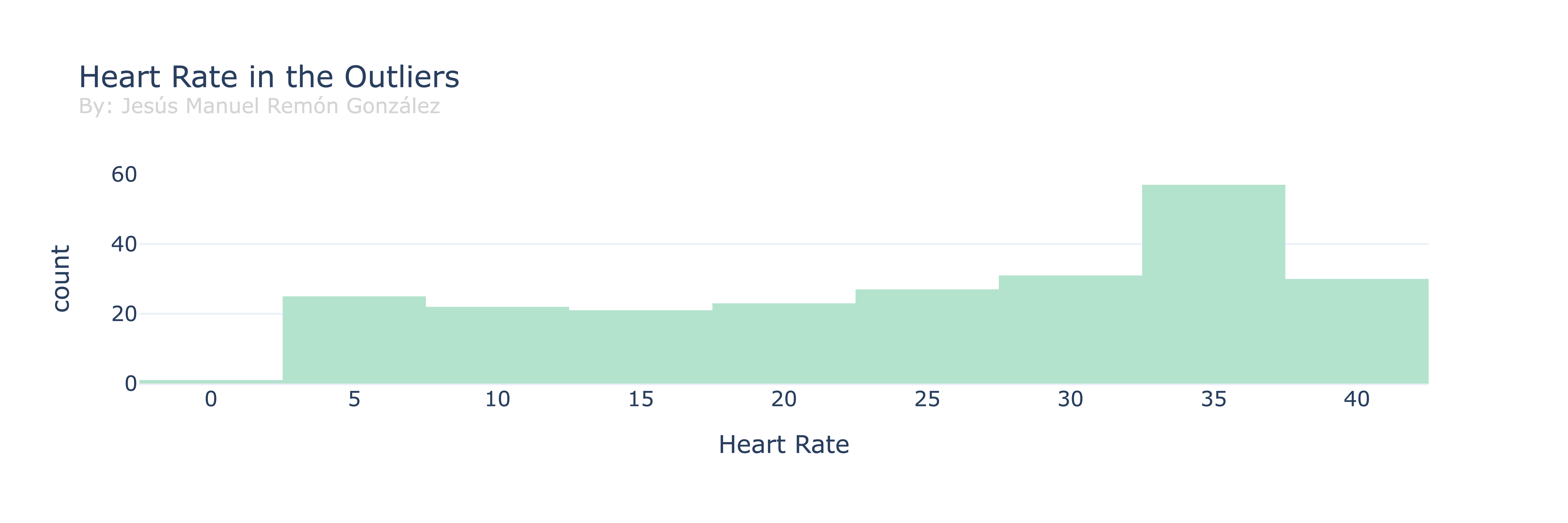
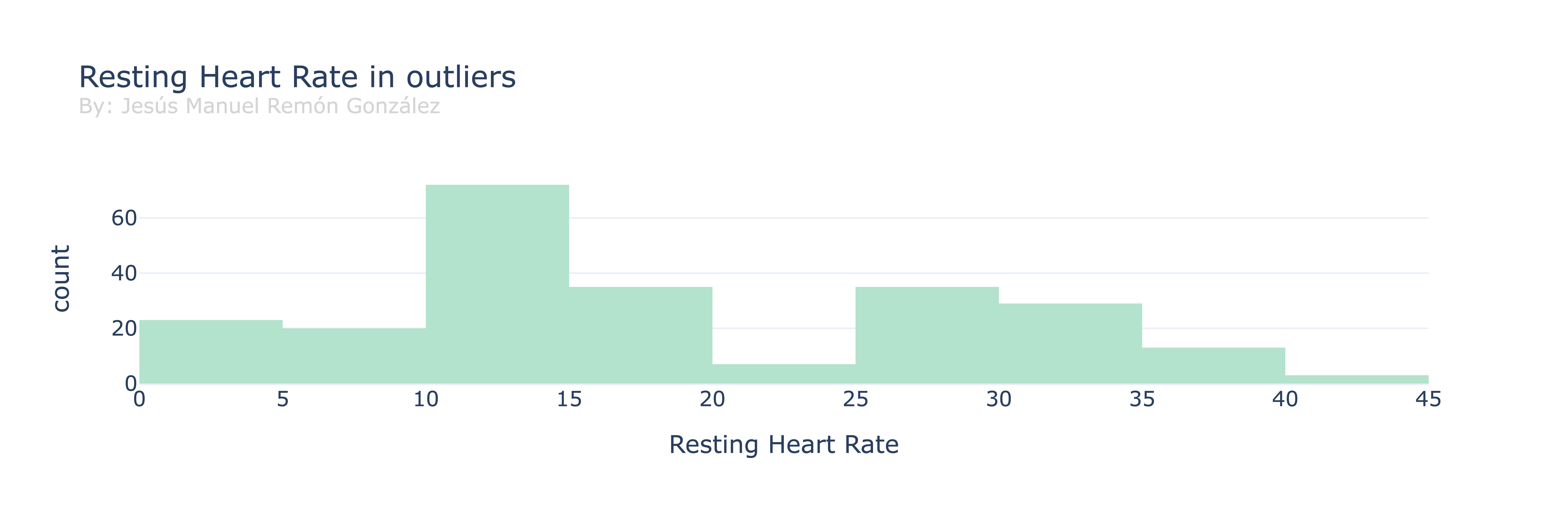
As we can see the cases where heart rate and resting heart rate is low are in the same group, so there could be a problem in using the device or a measurment error, we cannot know without having more information of the dataset.
Data Analysis
Height by Gender
With all the data clean, and after eliminating the outliers we can proceed to analyse the data and to investigate correlations and trends in the data.
Before starting to facility the easiness to read the information, I will replace in the gender column, the source does not indicate which one is for male and which one for female, so i will use the assumption that the height is bigger in the male case than for females.
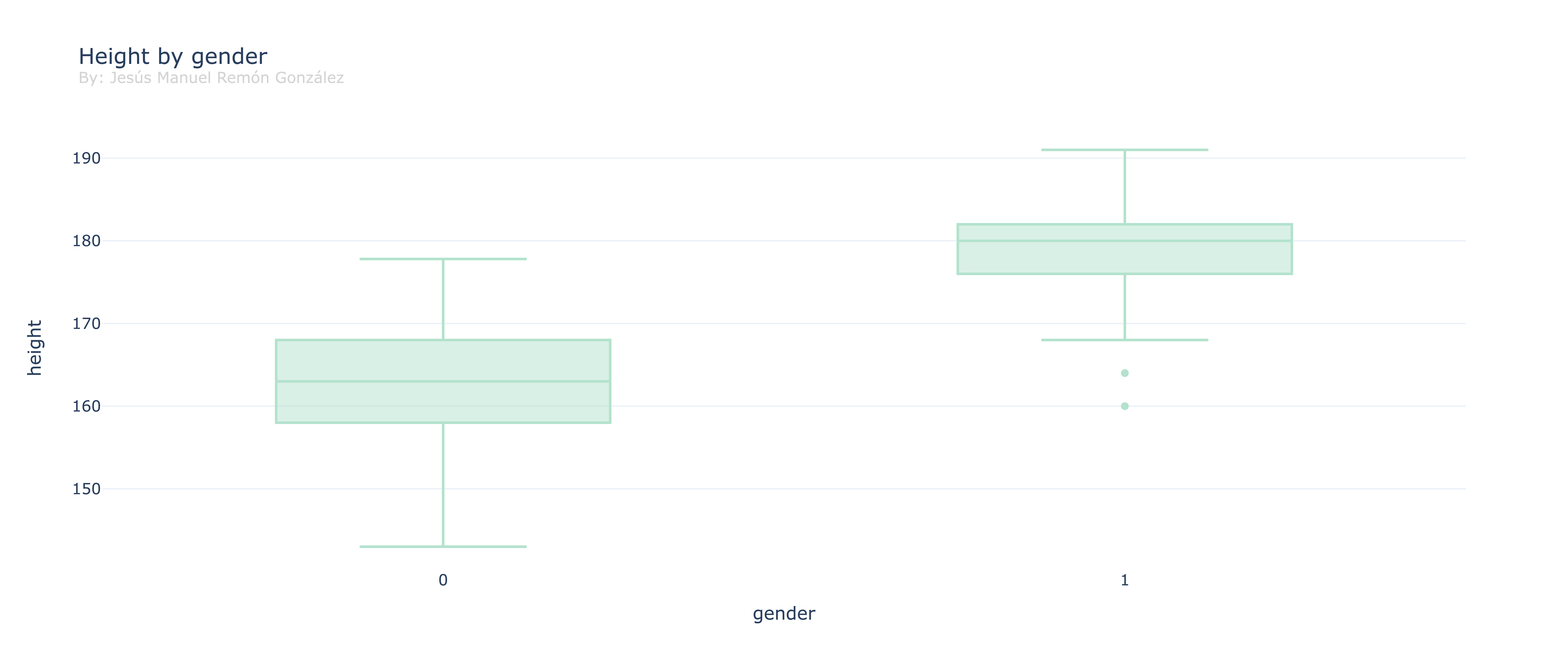
We can see that the 0 is lower than the 1, so we will use 0 for Female and 1 for Male.
health.gender = health.gender.replace({0: "Female", 1: "Male"})
Weight by Gender
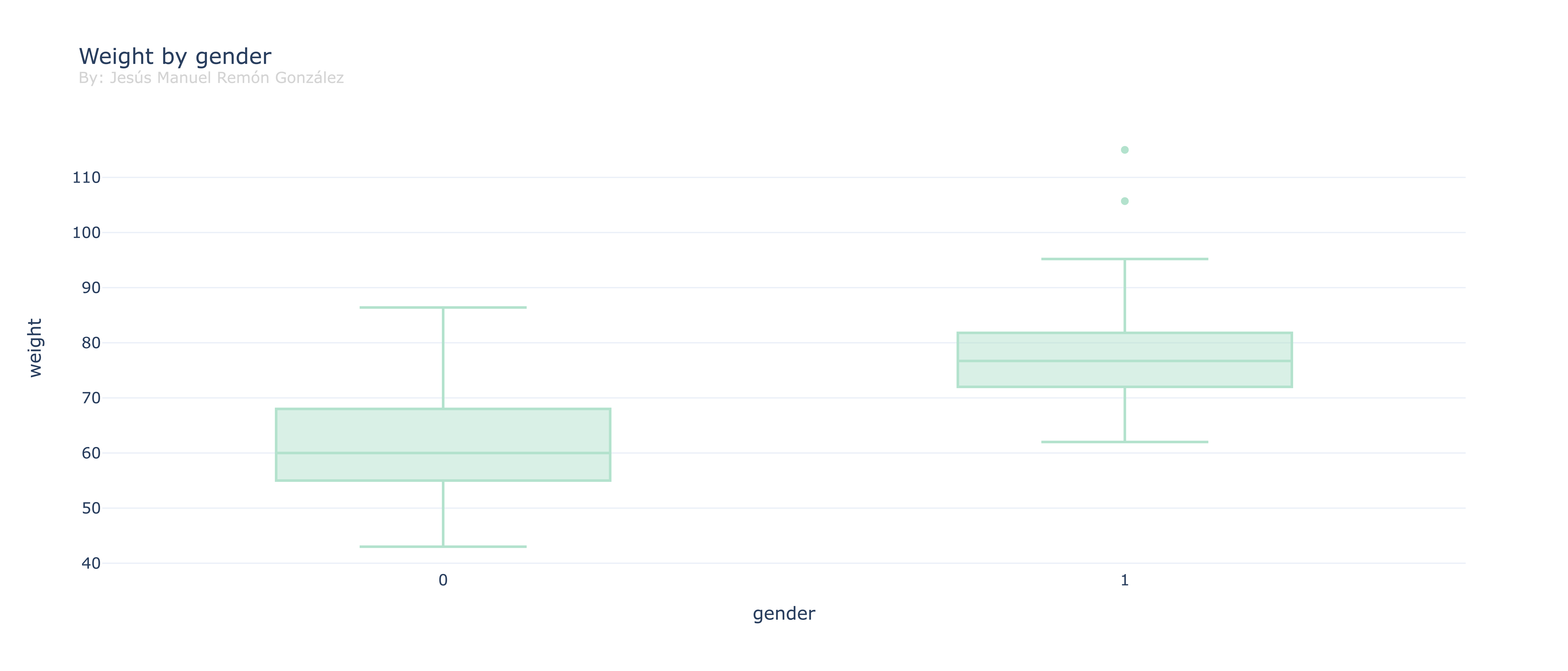
There is a deviation, where male present a higher weight compare to women in average, that is caused by a correlation between height and weight, even though we can find outliers, higher height usually implies higher mass. We can check that visualy by using a scatter plot.
Weight Vs Height
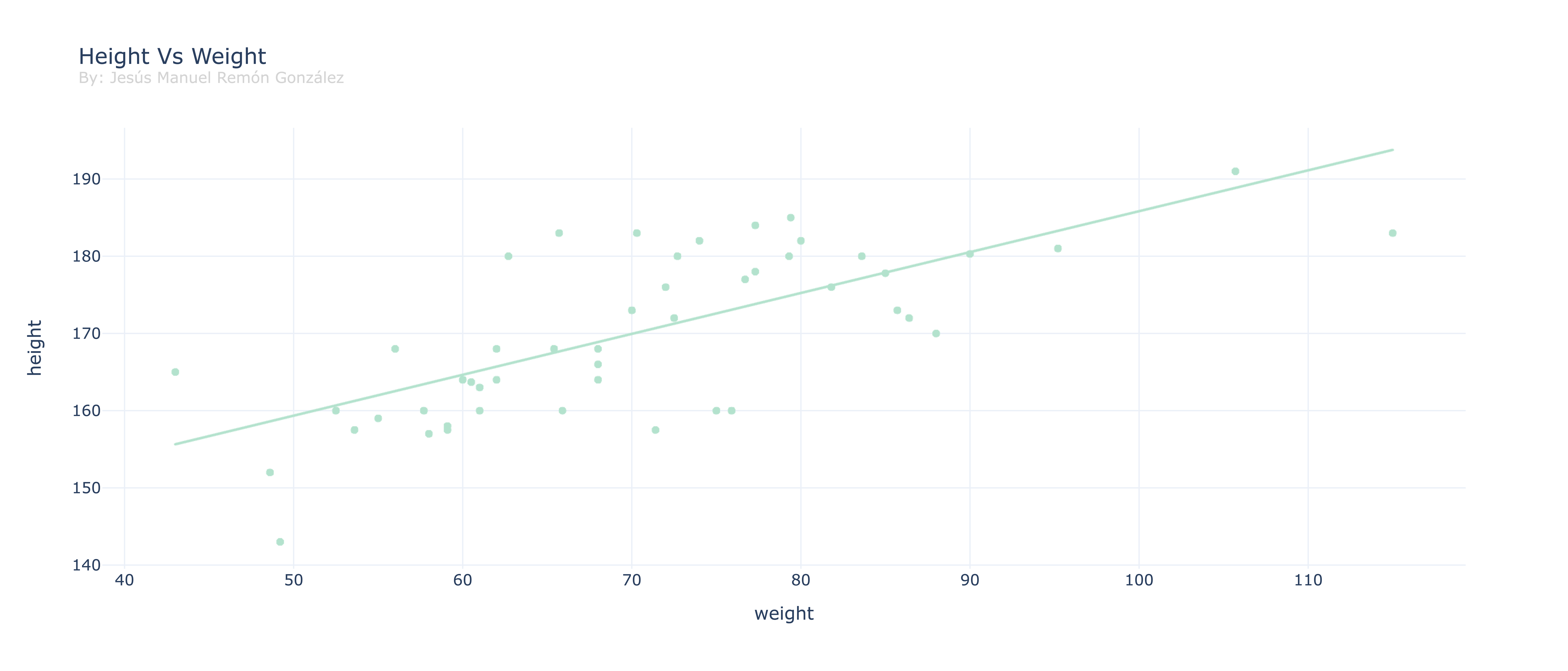
As we can see, the taller the person, the bigger is the weight, there are some outliers, as should be expected, but the trendline is a positive one.
Device by Gender
For this, let’s generate two new dataframes to differenciate the devices.
fitbit = health[health['device'] == 'fitbit']
fitbit_gender = fitbit.gender.value_counts()
| Gender | Count |
|---|---|
| Female | 1214 |
| Male | 1174 |
apple_watch = health[health['device'] == 'apple watch']
gender_apple = apple_watch.gender.value_counts()
| Gender | Count |
|---|---|
| Female | 1925 |
| Male | 1714 |
We can see that there is in general more females in both categories, there is no apparent differece, we can compare it by seeing the porportion of female vs male in the dataframe and then for each device.
| Pct. deviation Female Vs Male | Count |
|---|---|
| Total | 8.69% |
| Apple Watch | 12.31% |
| Fitbit | 3.41% |
We can see a bigger a difference in the proportion of women in the apple watch compare to the total of the dataset, that indicates that in our dataframe, there are a bigger proportion of women using apple watch, we cannot extrapolate any trend outside the dataframe.
Calories Burned by Device
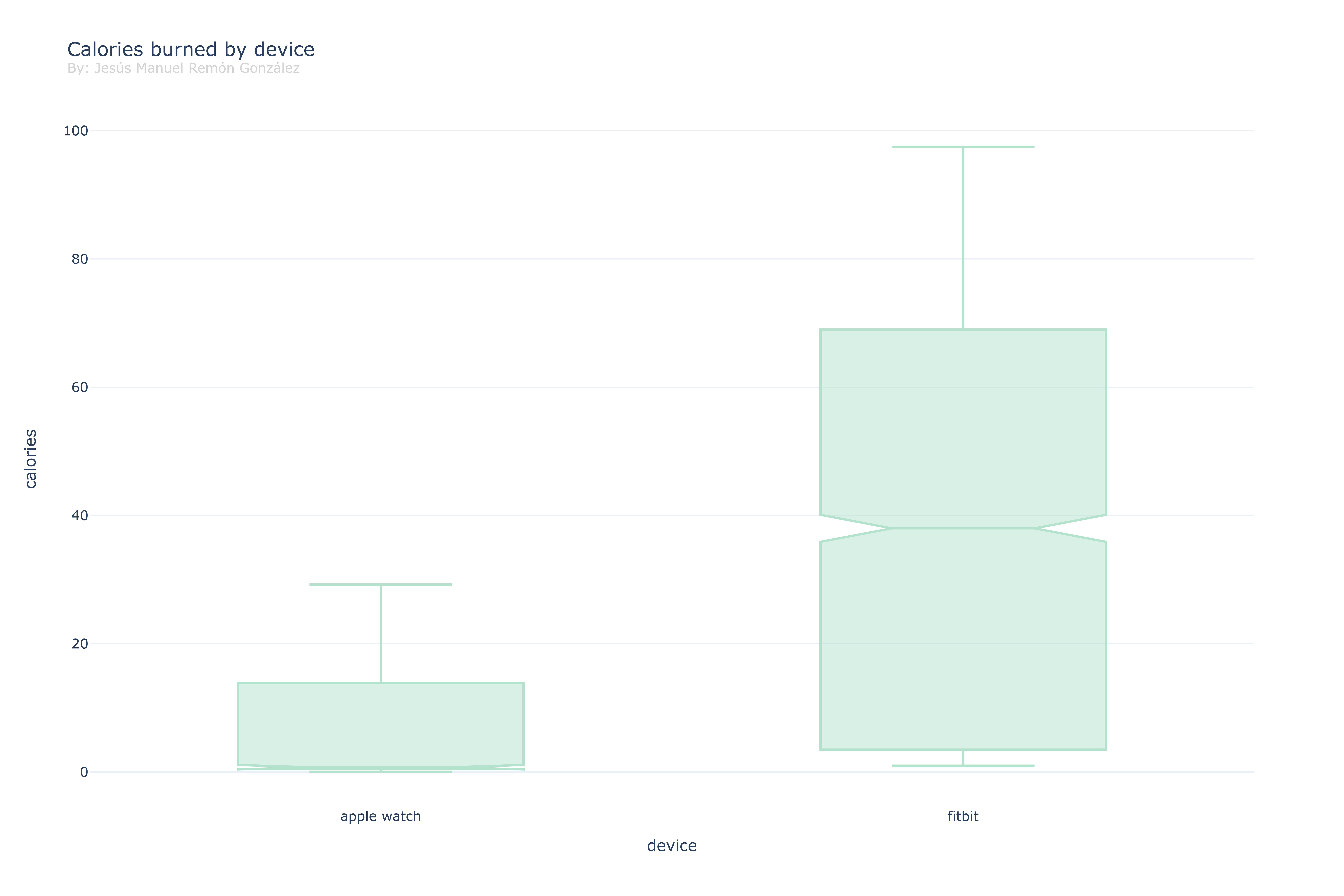
This shows a clear skew, the calories burned in the fitbit are higher, this may indicate that people using fitbit tend to perform more intense activities compare to the apple watch users that may use the watch not only for sport but also in a day to day basis to improve workflow.
Calories Burned by Activity
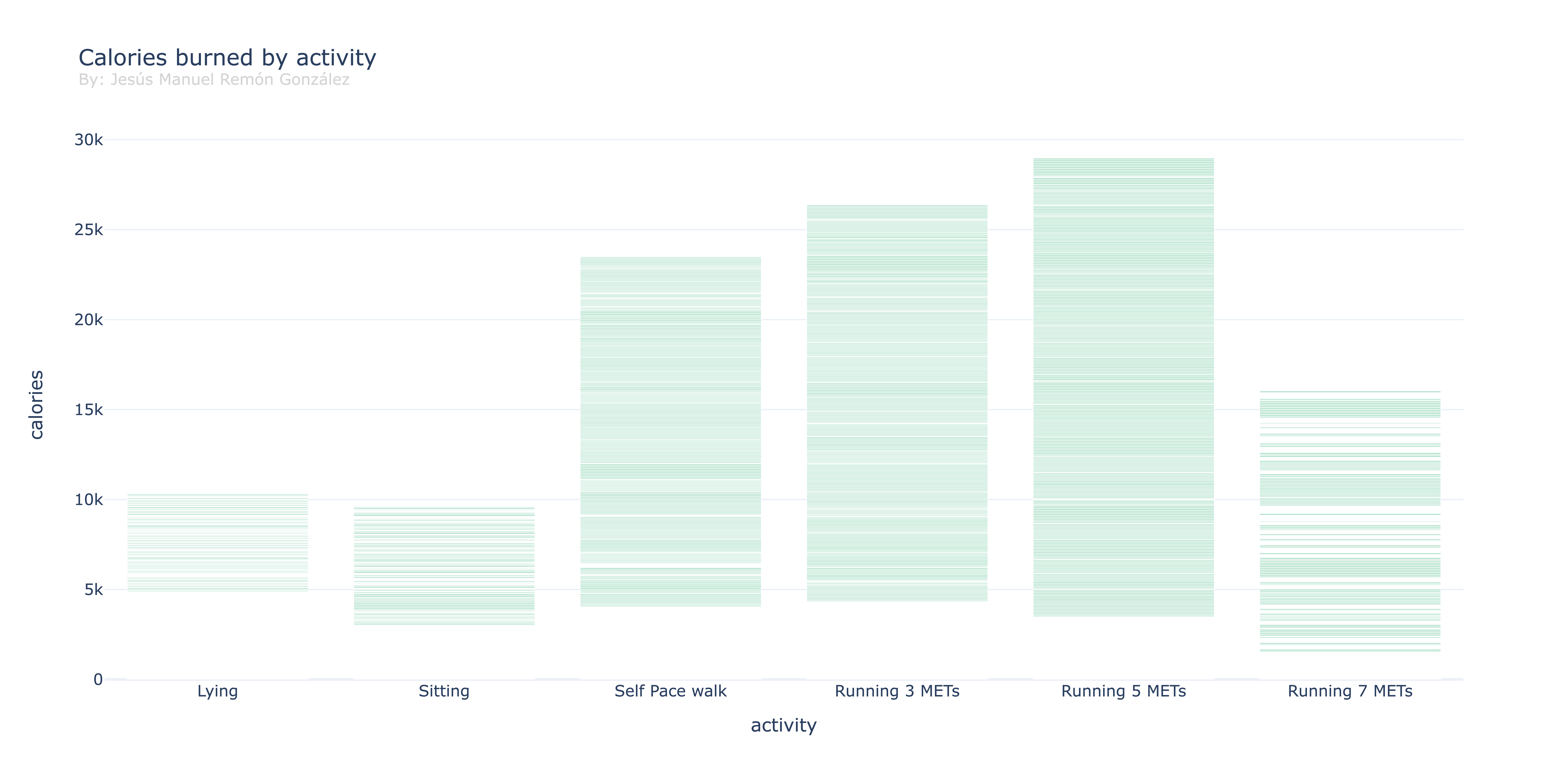
We can see that in general, the total amount of the mebers of the Dataframe add in total more calories burned by runing 5 METs, and that they count less in sitting than Lying, we can check which activity on average burned more calories, but we have to undertand that there is a lot of factor affecting how many calories you burned.
Heart Rate by Gender
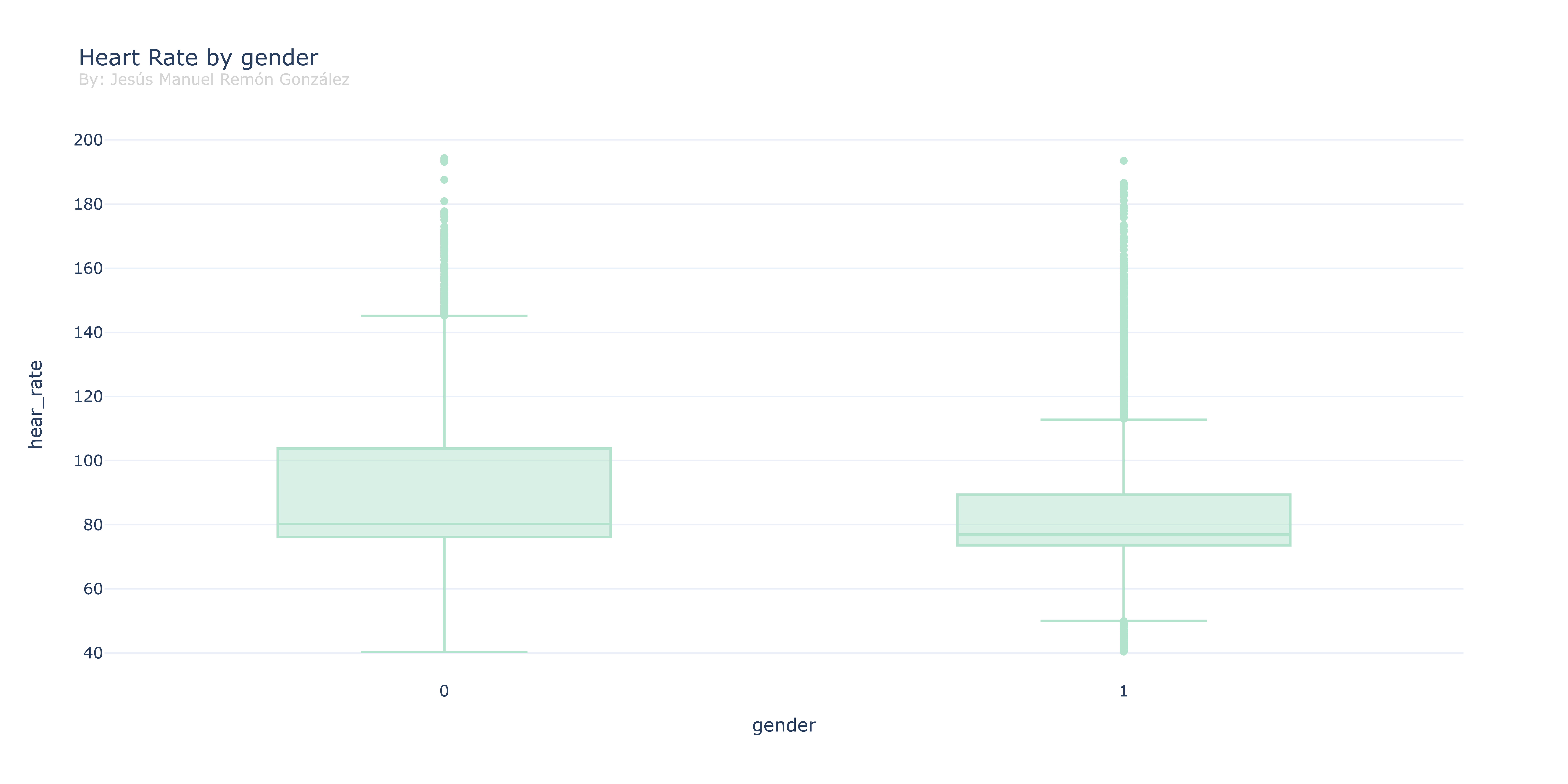
The graph reperesents a higher amplitude in the data for female comapre to men, with a higher median.
Steps by Gender
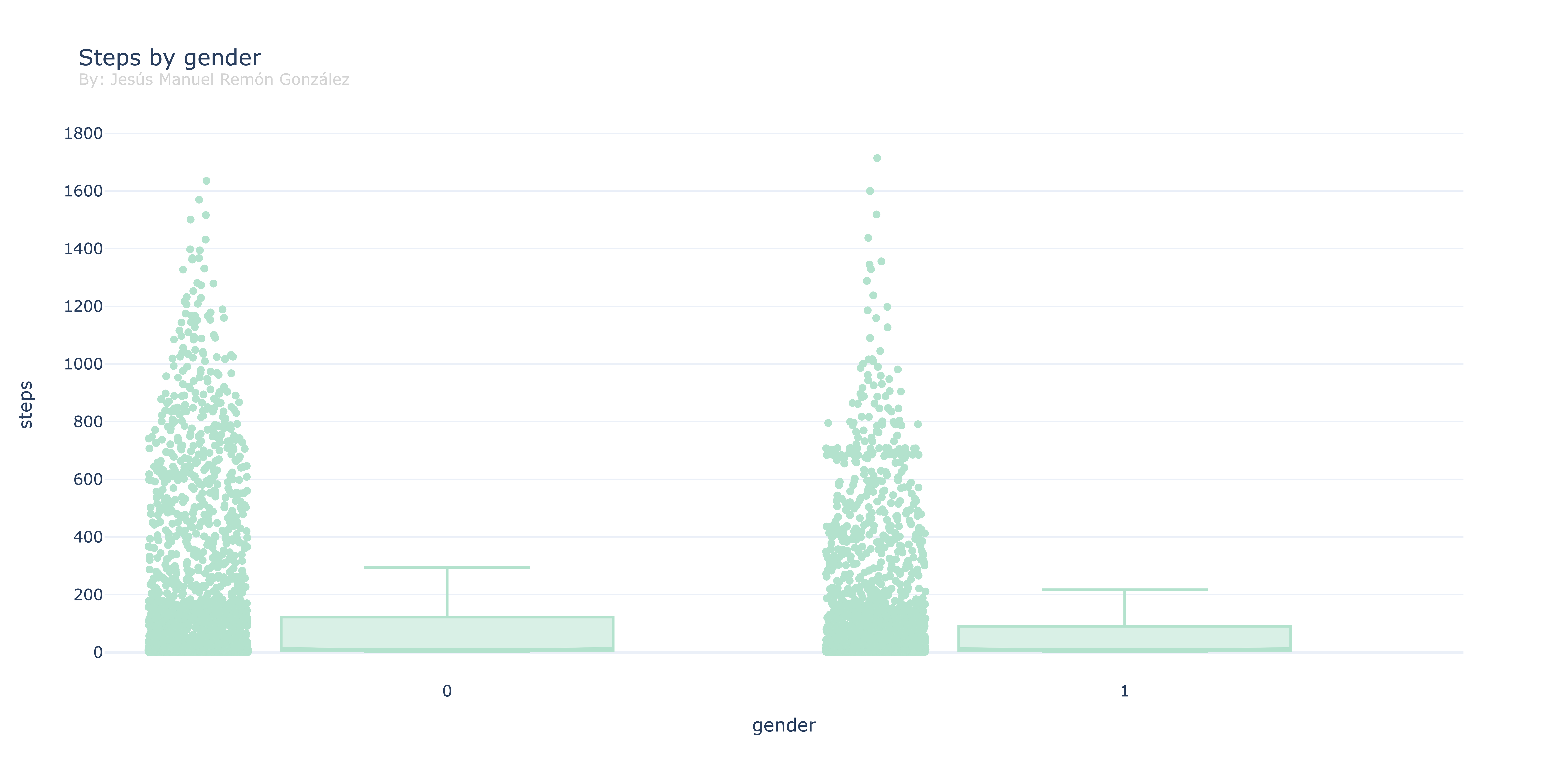
In this past case I have displayed the points to have a better representation of the data.
We can see that there is not a lot of difference between both gender, where female have a higher amount of steps in general, but a lower median compare to mean.
Steps by Device
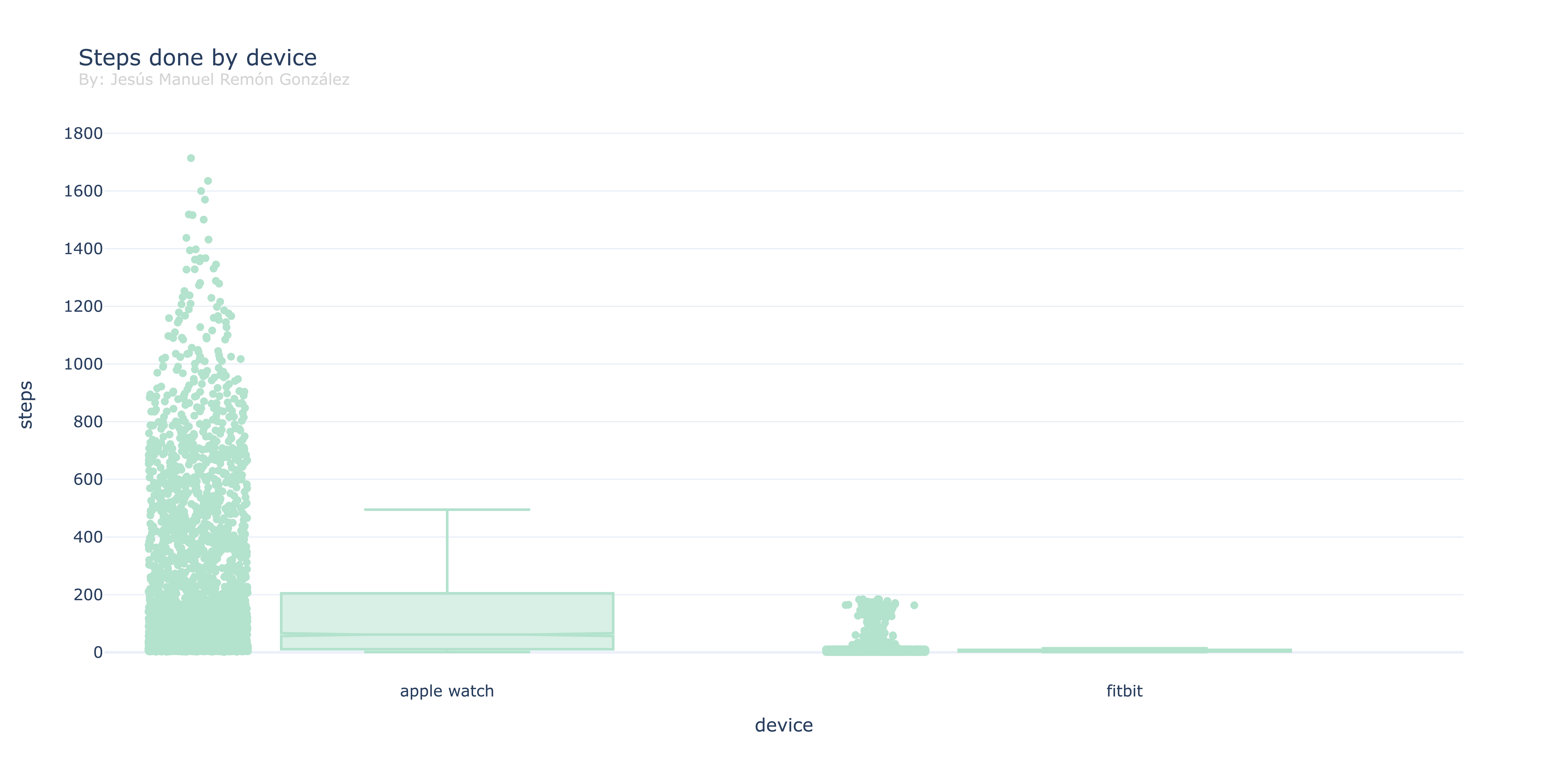
We can observe a big difference between the apple watch compared to the fitbit, this can be due to a different factors, such a big difference can indicate, a problem recording the steps.
Model Creation
Now that we all this data we can create a model to predict the calories burned with all the data we have, to create this model I will discard some of the columns, to keep it simpler, as this is for demostration and observation pouposes not for prediction accuracy.
First we will create a subset of the columns to use.
model_columns = ['age', 'gender', 'height', 'weight', 'steps', 'hear_rate', 'distance', 'resting_heart', 'device', 'activity', 'calories']
model_data = health[model_columns]
The next step is to convert the categorical data to integers to train the model.
model_data["gender"] = model_data["gender"].map({"Male": 1, "Female": 0})
model_data["device"] = model_data["device"].map({"apple watch": 0, "fitbit": 1})
model_data["activity"] = model_data["activity"].map({"Lying": 0, "Running 7 METs": 1, "Running 5 METs": 2, "Running 3 METs": 3, "Sitting": 4, "Self Pace walk": 5})
After that we can proceed to train the model.
x = np.array(model_data[['age', 'gender', 'height', 'weight', 'steps', 'hear_rate', 'distance', 'resting_heart', 'device', 'activity']])
y = np.array(model_data[['calories']])
xtrain, xtest, ytrain, ytest = train_test_split(x, y, test_size=0.10, random_state=42)
model = SVR()
model.fit(xtrain, ytrain)
Model Testing
After creating the model we could try to make a prediction using it. Is important to remember that we mapped the categorical values, so when we introduce the data we would need to encode using the same system.
Example Case: 29 year old male, that is 160 cm, with a weight of 80 kg, 2100 steps, with a heart rate of 90 bpms, a distance of 1000m, and a resting heart rate of 60 bpms. The device is an Apple Watch and the activity is running 7 mets.
features = np.array([[29, 1, 160, 80, 2100, 90, 1000, 60, 0, 1]])
print(model.predict(features))