In this Python-based project, I will study breast cancer patient care by predicting post-surgery survival rates. By analyzing crucial factors such as cancer type, stage, and age, my advanced machine learning models empower medical professionals to make informed decisions, optimize treatments, and improve patient outcomes in the fight against breast cancer.
Disclaimer This project is purely based on a sample dataset and is intended for educational and exploratory purposes only. It aims to facilitate data analysis and raise awareness about breast cancer. Please note that I am not a medical professional, and the outputs generated by this project should not be considered as medical advice. For any medical concerns or decisions related to breast cancer diagnosis, treatment, or care, it is crucial to consult with a qualified medical provider or a healthcare professional. They can provide personalized guidance and make informed decisions based on your specific medical history and needs.
Dataset Introduction
In this project, I will create an estimator for Breast Cancer, based on a dataset.
This dataset is composed of 400 breast cancer patients, and the data is available on Kaggle. The patients had surgery to remove the tumor, and the end column shows the survival status of each patient.
The columns are:
- Patient ID: ID of the patient.
- Age: Age of the patient.
- Gender: Gender of the Patient.
- Protein 1, 2, 3, and 4: Protein Expression Levels.
- Tumor Stage: Breast cancer stage.
- Histology: Infiltrating Ductal Carcinoma, Infiltrating Lobular Carcinoma, Mucinous Carcinoma.
- ER Status: Positive/Negative.
- PR Status: Positive/Negative.
- HER2 Status: Positive or negative.
- Surgery Type: Lumpectomy, Simple Mastectomy, Modified Radical Mastectomy, Other.
- Date of Surgery: The date of Surgery.
- Date of the last visit: The date of the last visit of the patient.
- Patient Status: Alive/Dead.
Notes
Some important notes before we start with the analysis. We will define some of the concepts in the dataset, using information provided by the American Cancer Society. For more information, refer to this link.
HER2 Status
HER2 is a protein that makes cancer cells grow quickly. Cancer cells with higher levels of HER2 are called HER2 Positive. They grow more quickly than HER2 negative cells but are also more likely to respond to drugs that target the HER2 protein.
The most common test for this is IHC (immunohistochemistry), but there are more options. Based on the IHC result, we can determine if the tumor is HER2 positive or negative.
- If the result is 0: HER2-Negative, no response to drugs that target HER2.
- If the result is 1+: HER2-Negative, usually no response to drugs that target HER2.
- If the result is 2+: Equivocal, because we don’t know, we need to perform another test, the FISH (fluorescence in situ hybridization).
- If the result is 3+: HER2-Positive, these tumors are usually treated with drugs that target HER2 protein.
After analyzing the HER2 Status, we will explain the two other columns, the ER status, and the PR Status, which are the Estrogen Receptor and the Progesterone Receptor, respectively.
ER Status and PR Status
Receptors are proteins on cells that can attach to certain substances in the blood. Normal breast cells and some breast cancer cells have receptors that attach to the hormones estrogen and progesterone and need these hormones for the cells to grow.
- ER Positive (ER+): Cancers that have strong receptors.
- PR Positive (PR+): Cancers that have progesterone receptors.
If the cancer cells have one or both of these receptors, it is called Hormone receptor positive (HR+). There are drugs that help prevent hormones (Estrogen and Progesterone) from attaching.
Whats the impact? Breast cancer that is hormone positive can be treated with hormone therapy drugs that lower estrogen levels or block estrogen receptors. Hormone receptor-positive cancers tend to grow more slowly than those that are hormone receptor-negative. Women with hormone receptor-positive cancers tend to have a better outlook in the short term, but these cancers can sometimes come back many years after treatment.
Project Code
Introduction
The first thing to do is establish the different libraries to import for the project. We will use pandas to work with the DataFrames, plotly to draw graphs to understand the project better, and Sklearn for model development.
import pandas as pd
import numpy as np
import plotly.express as px
from sklearn.model_selection import train_test_split
from sklearn.svm import SVC
Now that we have our libraries imported, we can proceed to open the data in pandas.
med_data = pd.read_csv('breast_cancer_survival.csv')
med_data.head()
| Age | Gender | Tumour_Stage | Histology | Surgery_type | Patient_Status |
|---|---|---|---|---|---|
| 42 | FEMALE | II | Infiltrating Ductal Carcinoma | Other | Alive |
| 54 | FEMALE | II | Infiltrating Ductal Carcinoma | Other | Dead |
| 63 | FEMALE | II | Infiltrating Ductal Carcinoma | Lumpectomy | Alive |
| 78 | FEMALE | I | Infiltrating Ductal Carcinoma | Other | Alive |
| 42 | FEMALE | II | Infiltrating Ductal Carcinoma | Lumpectomy | Alive |
This is an example of some of the columns and some of the rows (patients) of the DataFrame.
Cleaning Data
Before we proceed to analyze the data, we are going to clean it. The first thing is to analyze the null data in the dataset.
med_data.isnull().sum()
| Parameter | Null Values | Parameter | Null Values | |
|---|---|---|---|---|
| Age | 0 | Protein1 | 0 | |
| Protein2 | 0 | Protein3 | 0 | |
| Protein4 | 0 | Tumour_Stage | 0 | |
| Histology | 0 | ER status | 0 | |
| PR status | 0 | HER2 status | 0 | |
| Surgery_type | 0 | Date_of_Surgery | 0 | |
| Date_of_Last_Visit | 0 | Patient_Status | 0 | |
| Gender | 0 |
We don’t have any null values in the DataFrame. If we had any, there are multiple ways of resolving the issue, from more advanced to the simplest one. The most common would be to use the dropna() method to delete them.
med_data = med_data.dropna()
Once we have done that, we can start analyzing some of the information that we have. For example, the gender. Breast cancer can affect men and women, but it is more common in women.
Data Exploration
med_data.Gender.value_counts()
| Gender | |
|---|---|
| FEMALE | 313 |
| MALE | 4 |
We see a higher amount of female patients than male patients in the dataset, which makes sense since breast cancer is more common in women. However, the number of male patients is too small to draw any meaningful conclusions for this model.
Before training the model, we can see the difference in surgeries that the patients underwent and the percentage in the population sample. In the official dataset, there are three different surgeries and a category called other. The dataset owner does not specify the full content of the category, so we will use it for all the rest of surgeries not specified.
With that said, bfore proceding with the rest of the analysis, we can check the MALE data as is small.
gender_survival = med_data[med_data['Gender']=='MALE'].groupby('Gender')
| ID | Age | Gender | Tumour_Stage | Histology | Surgery_type | Patient_Status |
|---|---|---|---|---|---|---|
| 48 | 51 | MALE | II | Infiltrating Lobular Carcinoma | Simple Mastectomy | Alive |
| 143 | 44 | MALE | II | Infiltrating Lobular Carcinoma | Other | Dead |
| 191 | 68 | MALE | II | Infiltrating Ductal Carcinoma | Modified Radical Mastectomy | Alive |
| 322 | 84 | MALE | III | Infiltrating Ductal Carcinoma | Modified Radical Mastectomy | Alive |
The amount of data for MALE is to low to get a conclusion of it. But as a side note, we can see that 3 out of 4 survived, and that 3 out 4 had a tumour stage of II, and the other one III. All with the same Histology Infiltrating Lobular Carcinoma.
Infiltrating Lobular Carcinoma: Invasive lobular carcinoma, also known as infiltrating lobular carcinoma, begins in the milk-producing glands (lobules) of the breast. As an invasive type of cancer, ILC has spread beyond its original tumor site. Over time, ILC may become metastatic breast cancer.
stage_graph = med_data["Tumour_Stage"].value_counts()
stage = stage_graph.index
amount = stage_graph.values
figure = px.bar(data, x=stage, y=amount, title="% Of patients per Stage")
figure.show()
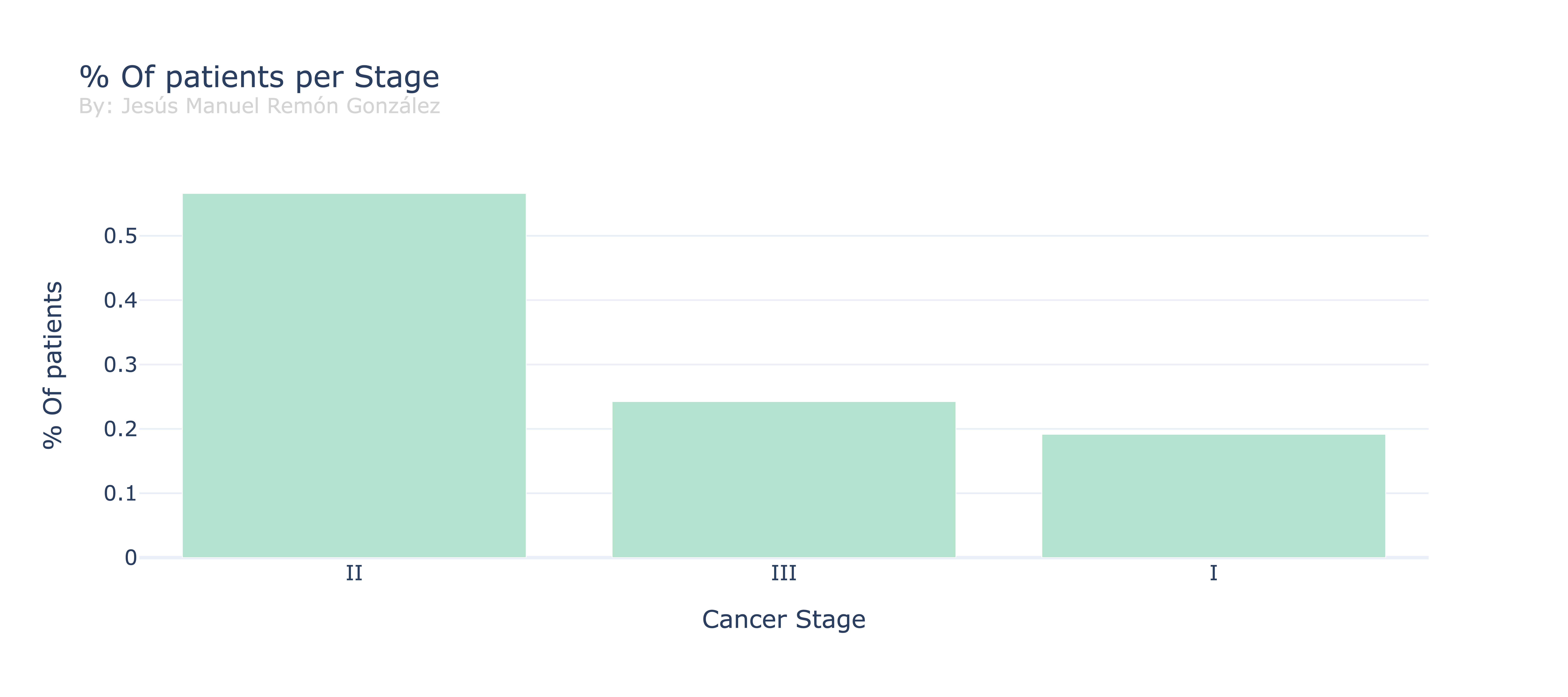
We see a higher amount of patients with stage II cancer compare to the others, so we have more information and, therefore more precission, for this model.
Before training the model, we can see a difference of the surgeries that the patient can get and the percentage in the population sample. In the official dataset there are three different sugeries and a category called other, the dataset owner does not specify the full content of the category, so we will use that for all the rest of surgeries non specify.
surgery_graph = med_data["Surgery_type"].value_counts()
surgery = stage_graph.index
amount = stage_graph.values
figure = px.pie(data, names=surgery, values=amount ,hole = 0.5,
title="Type of Surgery")
figure.show()
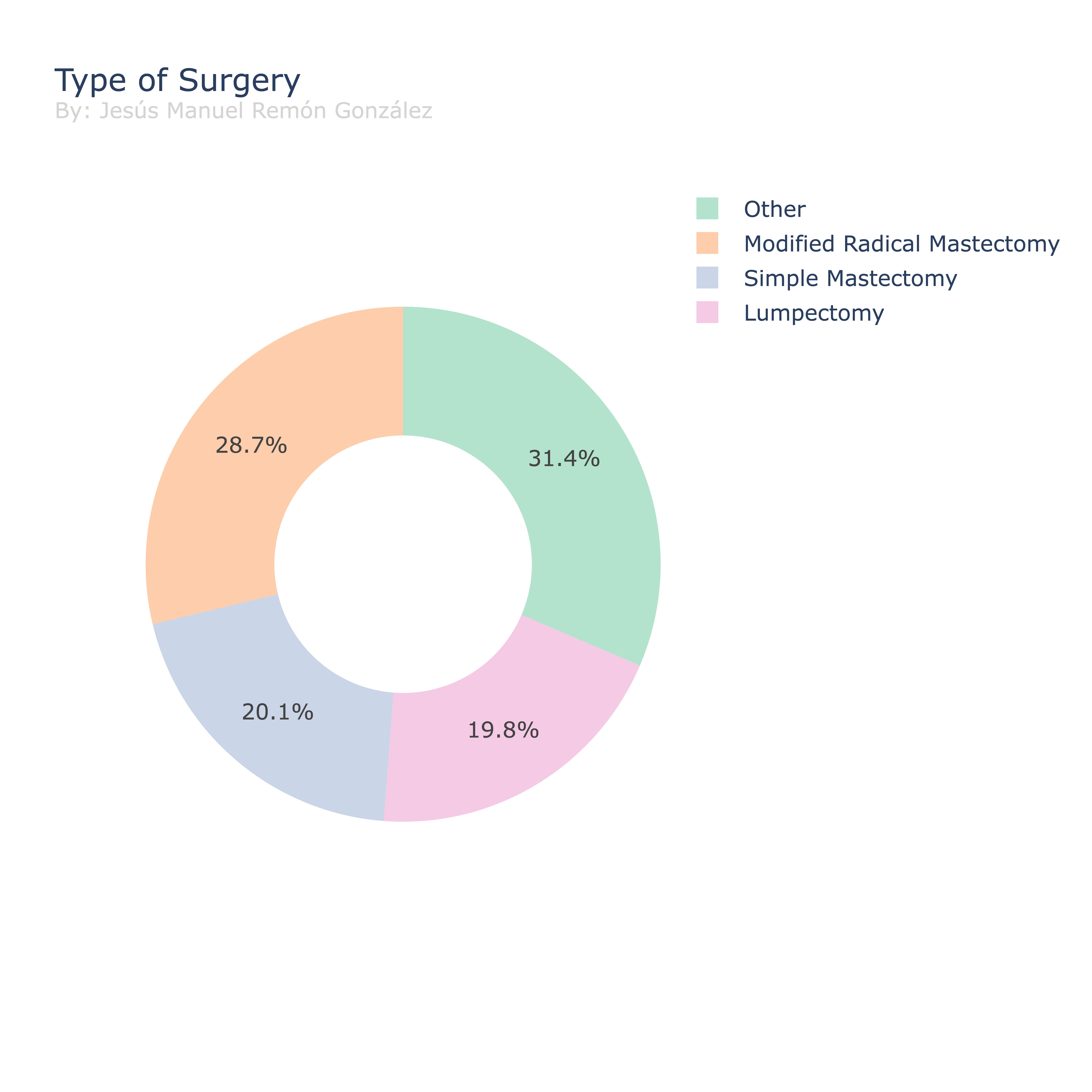
Data Analysis
Now that we understand the data and have performed some exploratory analysis, we can start analyzing some correlations in the data.
Hormone Status Postive Outlook
In the introduction of the porject, in the Notes section we talked about the impact of Hormone receptor-positive and its influence on tumor development. We will take a look at the data.
Seeing the dataframe, all the patients are double hormone positive (HR+) for Strogen and Progesterone but not all for HER2 protein status, lets check the different survival depending on the status of the HER2 protein.
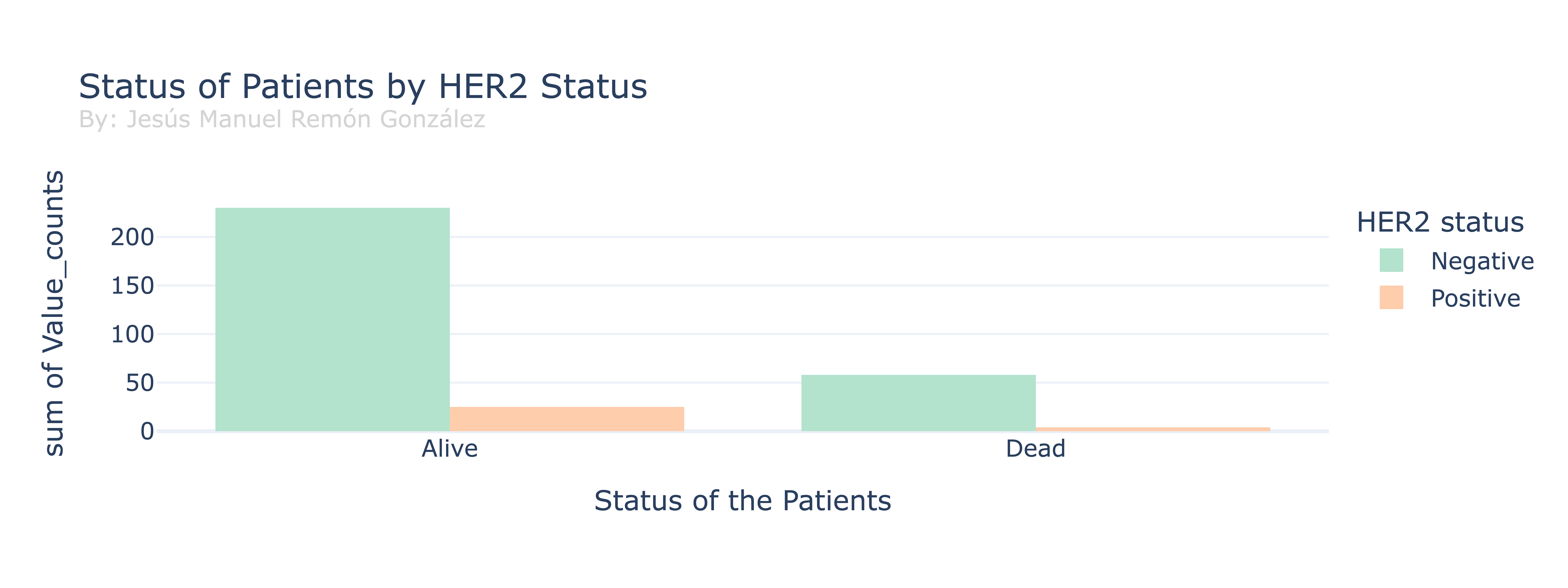
We can see a huge difference in the proportion of the status in the patients, inside the different status, there is also a huge difference in each group between the positives and negatives for HER2, that indicates us that the DataFrame have a lot more patients with a doble Hormone positive and a minority for triple positive.
Also the difference in patients with a HER2 status negative that are alive vs dead is bigger than for positive, but that can be cause by the fact that there is a bigger sample. According to the literature, we would porbably see a bigger amount of alive patients for HER2 positive status, as the studies suggest a more positive outcome in the short term for those patients.
Surgeries status
Now we will perform a comparison between the different types of surgeries and the status of the patients after the surgery.
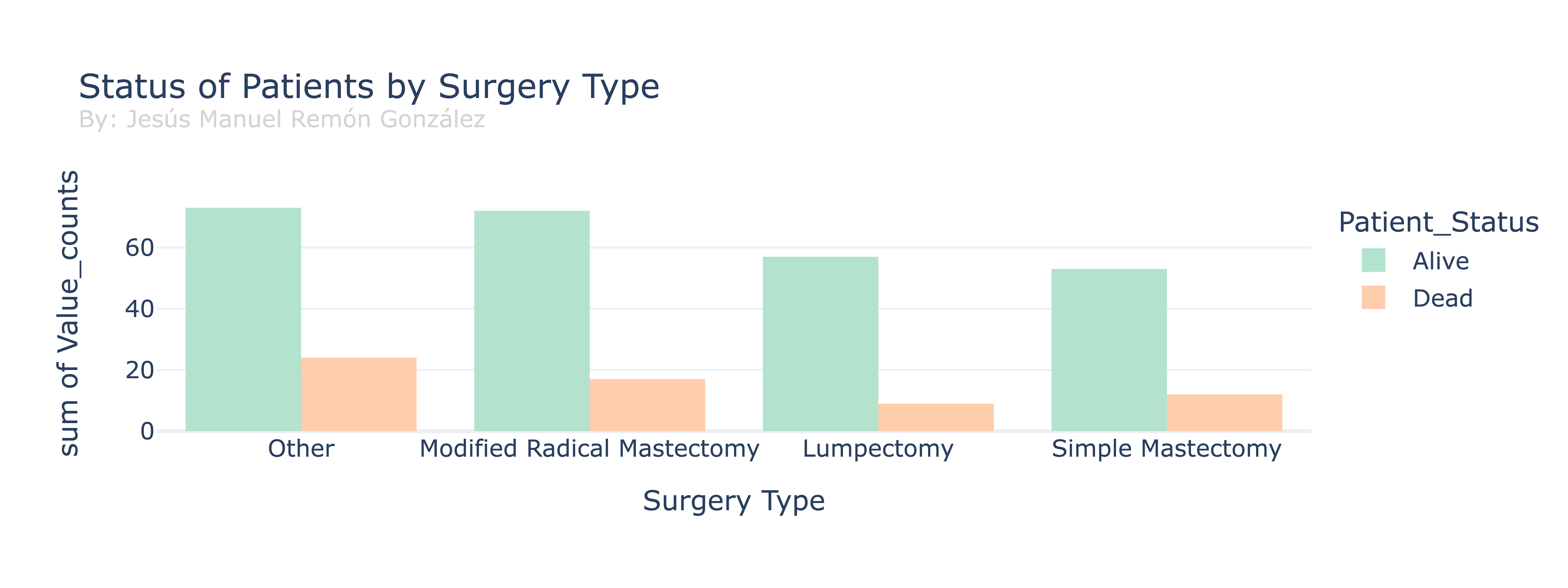
The difference in number in alive Vs dead patients in the dataframe can be due to the type of cancer or the patients, so without more data a conclussion cannot be draw from it. One interesting thing to see is the proportion of alive vs dead patients in each surgery group. The lumpectomy presents a higher proportion than the simple masectomy, can be cause by different factors, but of them that we can check is the stage for each surgery.
Lumpectomy: A lumpectomy is a breast surgery that involves removing a localized tumor while preserving most of the healthy breast tissue. It is commonly used for early-stage breast cancer treatment, aiming to achieve cancer removal with minimal impact on the breast’s appearance. Additional treatments like radiation therapy may follow to reduce the risk of cancer recurrence.
Based in the medical description we can see that the higher porportion may be cause by the fact that the Lumpectomy is usually used for early stages patients. Let’s check our DataFrame.
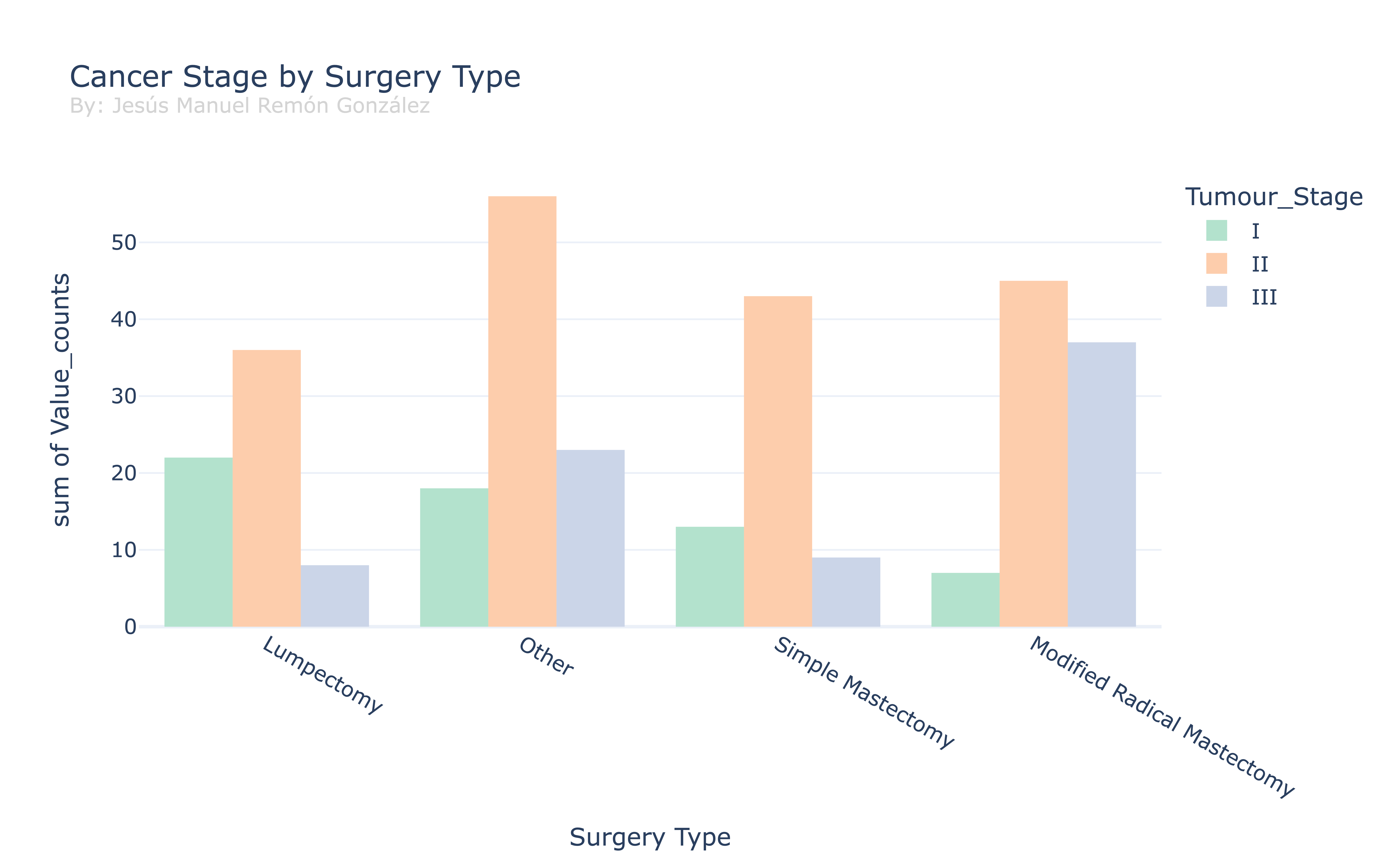
As we commented, the lumpectomy is done more in patients in stage I cancer, while the simple masectomy is more used in Stage II cancer.
Model Training
Once we have done some data analysis, we can proceed to model training. We need to be aware that many variables are categorical, so before continuing, we have to convert them.
med_data["Tumour_Stage"] = med_data["Tumour_Stage"].map({"I": 1, "II": 2, "III": 3})
med_data["Histology"] = med_data["Histology"].map({"Infiltrating Ductal Carcinoma": 1,
"Infiltrating Lobular Carcinoma": 2, "Mucinous Carcinoma": 3})
med_data["ER status"] = med_data["ER status"].map({"Positive": 1})
med_data["PR status"] = med_data["PR status"].map({"Positive": 1})
med_data["HER2 status"] = med_data["HER2 status"].map({"Positive": 1, "Negative": 2})
med_data["Gender"] = med_data["Gender"].map({"MALE": 0, "FEMALE": 1})
med_data["Surgery_type"] = med_data["Surgery_type"].map({"Other": 1,
"Modified Radical Mastectomy": 2, "Lumpectomy": 3, "Simple Mastectomy": 4})
We will also use this to make predictions for the model.
x = np.array(data[['Age', 'Gender', 'Protein1', 'Protein2', 'Protein3','Protein4',
'Tumour_Stage', 'Histology', 'ER status', 'PR status', 'HER2 status',
'Surgery_type']])
y = np.array(data[['Patient_Status']])
xtrain, xtest, ytrain, ytest = train_test_split(x, y, test_size=0.10, random_state=42)
model = SVC()
model.fit(xtrain, ytrain)
With this, we have finished the model and can proceed to test it with our own cases to see the output.
Model Usage
With the model trained, we can use it to predict the status of different patients. We have to remember that for the model, we have encoded the categorical values, so we will use the same encoding for the parameters. For the protein expressions, I will use the same values for both cases.
Case 1
The patient is a Female, 36 years Old, Stage III Tumour, the histology is Infiltrating Ductal Carcinoma, a triple positive hormone status, that means Progesterone, Strogen and HER2 positive, the surgery was a Modified Radical Mastectomy.
| Age | Gender | Tumour Stage | Histology | Surgery Type | Result |
|---|---|---|---|---|---|
| 36 | 1 | 3 | 1 | 2 | Alive |
Case 2
The patient is a Female, 25 years Old, Stage II Tumour, the histology is Infiltrating Lobular Carcinoma, a double positive hormone status, that means Progesterone, Strogen positive but HER2 negative, the surgery was a Lumpectomy.
| Age | Gender | Tumour Stage | Histology | Surgery Type | Result |
|---|---|---|---|---|---|
| 25 | 1 | 2 | 2 | 3 | Alive |